2010 - Transplantomics and Biomarkers in Transplantation
This page contains exclusive content for the member of the following sections: TTS. Log in to view.
Posters
10.15 - NOVEL CELL-TYPE SPECIFIC DECONVOLUTION OF WHOLE-BLOOD GENE EXPRESSION PROFILES IN RENAL ACUTE REJECT
Presenter: Purvesh, Khatri, Stanford, USA
Authors: Purvesh Khatri, Shai Shen-Orr, Robert Tibshirani, Atul Butte, Minnie Sarwal.
NOVEL CELL-TYPE SPECIFIC DECONVOLUTION OF WHOLE-BLOOD GENE EXPRESSION PROFILES IN RENAL ACUTE REJECTION
Purvesh Khatri, Shai Shen-Orr, Robert Tibshirani, Atul Butte, Minnie Sarwal. Department of Pediatrics, School of Medicine, Stanford University, Stanford, CA, USA.
Background: Although, a set of blood-based biomarkers have been identified for acute rejection, the expression profile of different blood cell-types in rejection is unknown.
Methods: We developed a novel statistical deconvolution method for identifying cell-type specific gene expression profiles using whole blood microarray expression data and relative frequencies of individual cell types in each sample. Whole blood gene expression data from 24 renal transplant patients were analyzed on Affymetrix HGU133 plus 2 whole genome expression arrays. Of the 24 patients, 15 samples were from biopsy-proven acute rejection patients and 9 were from stable patients. Simultaneous Complete Blood Counts (CBCs), containing relative frequencies of monocytes, lymphocytes, eosinophils, basophils, and neutrophils, were available for each sample.
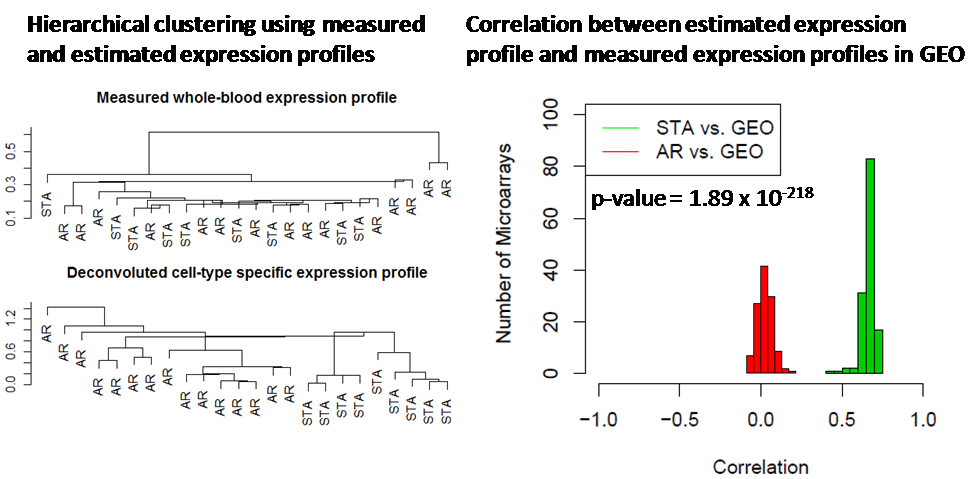
Results: Whole blood gene expression analysis using SAM did not identify any significantly differentially expressed genes between AR and STA patients at FDR of 10%. We estimated expression profiles for each cell type in each sample using statistical deconvolution. Significance analysis of the estimated expression profiles identified 213 genes significantly upregultaed in AR in one of the cell types at an FDR of 5%. Furthermore, while hierarchical clustering using whole blood expression data does not classify the samples correctly, the deconvoluted gene expression profile in specific cell subsets could distinctly cluster the samples in two groups (Fig. 1). In addition, we downloaded 132 microarrays of a specific blood cell type from NCBI GEO. As shown in Fig. 1, the estimated cell-specific expression profile in STA group is highly correlated with measured expression profiles in that cell type, whereas the estimated cell-specific expression profile in AR group does not correlate with the measured cell-specific expression profile, which clearly show that during rejection gene expression profiles of a specific cell-type are clearly disrupted .
Conclusion: Our novel statistical deconvolution method is able to identify the specific subset of blood cells that correlates with acute rejection in renal allografts.
Important Disclaimer
By viewing the material on this site you understand and accept that:
- The opinions and statements expressed on this site reflect the views of the author or authors and do not necessarily reflect those of The Transplantation Society and/or its Sections.
- The hosting of material on The Transplantation Society site does not signify endorsement of this material by The Transplantation Society and/or its Sections.
- The material is solely for educational purposes for qualified health care professionals.
- The Transplantation Society and/or its Sections are not liable for any decision made or action taken based on the information contained in the material on this site.
- The information cannot be used as a substitute for professional care.
- The information does not represent a standard of care.
- No physician-patient relationship is being established.
Contact
Address
The Transplantation Society
International Headquarters
740 Notre-Dame Ouest
Suite 1245
Montréal, QC, H3C 3X6
Canada
